Case-Study III: d-dimensional multimodal mixture distribution
To test the performance of DREAM in the presence of multi-modality, the third case study involves a d = 10-dimensional bimodal mixture of two Gaussian distributions
|
(3.01) |
where and -5 and 5 are d-dimensional vectors that define the mean of each normal distribution, and the covariance matrix is equivalent to the identity-matrix Id, a 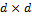 (square) matrix with ones on the main diagonal and zeros elsewhere. The density of the target distribution is simply the sum of the pdfs of two normal distributions
(square) matrix with ones on the main diagonal and zeros elsewhere. The density of the target distribution is simply the sum of the pdfs of two normal distributions
|
(3.02) |
This equation can be simplified as 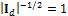 and the term
and the term 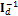 in the exponent can be removed without harm. This leads to
in the exponent can be removed without harm. This leads to
|
(3.03) |
The initial state of each Markov chain (= starting point) is drawn from 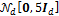 , the
, the 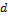 -variate normal distribution with mean zero and covariance matrix equal to a multiple (five) of the identity matrix. We use N = 20 different chains with DREAM and apply default values of the algorithmic variables.
-variate normal distribution with mean zero and covariance matrix equal to a multiple (five) of the identity matrix. We use N = 20 different chains with DREAM and apply default values of the algorithmic variables.
Implementation of plugin functions
The complete source code can be found in DREAM SDK - Examples\D1\Drm_D1_Example03\Plugin\Src_Cpp
///////////////////////////////////////////////////////////////////////////////////////////////////
// EvaluatorModel.cpp
///////////////////////////////////////////////////////////////////////////////////////////////////
//-------------------------------------------------------------------------------------------------
HRESULT STDMETHODCALLTYPE CEvaluatorExample03::GetInputParams
//-------------------------------------------------------------------------------------------------
(
IDreamInputParams* pIDreamInputParams // Pointer to DREAM input data to be defined
)
///////////////////////////////////////////////////////////////////////////////////////////////////
/*
DESCRIPTION:
The purpose of this function is to define default DREAM parameters for this particular project.
This function is called by the main program to initialize data of each newly created simulation
case. This data can be later modified in the GUI (except parameters locked by flag
DreamSolver::disable_edit) or can be overwritten while reading case data from a file.
In the AppTest console application, there is no GUI and this functions actually defines all
input parameters that are subsequently used by DREAM Solver.
RETURN VALUE:
S_OK - Operation successful
S_FALSE - Function not implemented (program will continue)
E_FAIL - Operation failed (program will stop)
*/
///////////////////////////////////////////////////////////////////////////////////////////////////
{
// Always check pIDreamInputParams pointer
if(pIDreamInputParams == nullptr)
{
return E_INVALIDARG;
}
// Set default input parameters
pIDreamInputParams->SetProblemDimension(10, DreamSolver::enable_edit);
pIDreamInputParams->SetNumberOfMarkovChains(pIDreamInputParams->GetProblemDimension(),
DreamSolver::enable_edit);
pIDreamInputParams->SetNumberOfGenerations(100000, DreamSolver::enable_edit);
pIDreamInputParams->SetThinnigSampleToStore(10, DreamSolver::enable_edit);
pIDreamInputParams->SetLikelihoodChoice(eLikelihood::LIK_102, DreamSolver::disable_edit);
pIDreamInputParams->SetInitialDistribution(eInitDistrib::eInitNormal, DreamSolver::enable_edit);
pIDreamInputParams->SetOutlierTest(eOutlierTest::eOutlierIqr, DreamSolver::enable_edit);
pIDreamInputParams->SetBoundHandling(eBoundHandling::eBoundUnbounded, DreamSolver::enable_edit);
// Example can run in parallel mode (recommended mode)
pIDreamInputParams->SetParallelMode(eParallelModeYes, DreamSolver::enable_edit);
return S_OK;
}
//-------------------------------------------------------------------------------------------------
HRESULT STDMETHODCALLTYPE CEvaluatorExample03::InitData
//-------------------------------------------------------------------------------------------------
(
IDreamPluginInfo* pIPluginInfo // Plugin information
, IDreamInputParViewer* pIDreamInputParams // Current DREAM input parameters
)
///////////////////////////////////////////////////////////////////////////////////////////////////
/*
DESCRIPTION:
This function is called to prepare:
A/ DREAM input data before accessing them via the IDreamDataSource interface and
B/ all other local data required by evaluator for the calculation.
It can be also used to read needed data from a file in directory pIPluginInfo->m_strModelDataDir
(directory ...Drm_ExampleXX\Model\Data).
Remark 1: This function prepares DREAM input data in the evaluator according to current
DREAM parameters defined by pIPluginInfo interface. Note that some parameters can be
different from default parameters defined in function CEvaluatorExample::GetInputParams().
Remark 2: This function should be called before using functions
CEvaluatorExample::GetMinMaxData, ::GetNormalData, ::GetPriorData, ::GetPriorDataCustom,
::GetMeasurementData and ::GetBayesData. However, function CEvaluatorExample::GetInputParams()
can be called independently.
RETURN VALUE:
S_OK - Operation successful
S_FALSE - Function not implemented (program will continue)
E_FAIL - Operation failed (program will stop)
*/
///////////////////////////////////////////////////////////////////////////////////////////////////
{
// Check parameters
if(pIPluginInfo == nullptr || pIDreamInputParams == nullptr)
{
// Invalid argument
return E_INVALIDARG;
}
// Free memory from previous calculation
FunctionsSC::DeleteMatrix(m_L, m_Lsize, m_Lsize);
FunctionsSC::DeleteMatrix(m_invL, m_invLsize, m_invLsize);
FunctionsSC::DeleteMatrix(m_mu, m_k, m_muSize);
FunctionsSC::DeleteVector(m_logF);
int d = pIDreamInputParams->GetProblemDimension();
// How many Gaussians?
m_k = 2;
// Determine weight of each respective Gaussian
// log_prior = log ( [1/3 2/3] );
double* logPrior = nullptr;
FunctionsSC::CreateVector(logPrior, m_k);
logPrior[0] = log(1. / 3.);
logPrior[1] = log(2. / 3.);
// Mean of both modes
//mu = [ -5*ones(1,d) ; 5*ones(1,d) ];
m_muSize = d;
double* mu1 = nullptr;
double* mu2 = nullptr;
FunctionsSC::CreateVector(mu1, d, -5.0);
FunctionsSC::CreateVector(mu2, d, 5.0);
FunctionsSC::CreateMatrix(m_mu, m_k, d);
FunctionsSC::MatrixSetRow(0, mu1, m_mu, m_k, d);
FunctionsSC::MatrixSetRow(1, mu2, m_mu, m_k, d);
// Now compute the inverse of the covariance matrix
double** C = nullptr;
FunctionsSC::IdentityMatrix(C, d, d);
// compute the log determinant of covariance
m_Lsize = d;
if(!FunctionsSC::Cholesky(m_L, C, d))
{
return E_FAIL;
}
m_invLsize = d;
FunctionsSC::CopyMatrix(m_invL, m_L, d);
if(!FunctionsSC::Inv(m_invL, d))
{
return E_FAIL;
}
double* diagL = nullptr;
FunctionsSC::MatrixDiagonal(diagL, m_L, d);
// Calculate part of normalization constant
// logDetSigma = 2 * sum ( log ( diagL ) );
FunctionsSC::VectorElementLog(diagL, d);
double logDetSigma = 2.0 * FunctionsSC::VectorElementSum(diagL, d);
// Calculate total normalization constant (k values)
// m_log_F = -0.5 * logDetSigma + logPrior - d * log(2 * pi) / 2;
double value = -0.5 * logDetSigma - d * log(2 * pi<double>()) / 2.0;
double* vector2 = nullptr;
FunctionsSC::CreateVector(vector2, m_k, value);
FunctionsSC::VectorVectorAddition(m_logF, logPrior, m_k, vector2, m_k);
FunctionsSC::DeleteVector(mu1);
FunctionsSC::DeleteVector(mu2);
FunctionsSC::DeleteVector(logPrior);
FunctionsSC::DeleteVector(diagL);
FunctionsSC::DeleteVector(vector2);
FunctionsSC::DeleteMatrix(C, d, d);
return S_OK;
}
//-------------------------------------------------------------------------------------------------
HRESULT STDMETHODCALLTYPE CEvaluatorExample03::GetMinMaxData
//-------------------------------------------------------------------------------------------------
(
IDreamInputParViewer* pInputPar // Interface to DREAM input parameters
, IDreamDataMinMax* pIMinMaxData // Interface to Min/Max data
)
///////////////////////////////////////////////////////////////////////////////////////////////////
{
// Always check pIMinMaxData pointer
if(pIMinMaxData == nullptr)
{
return E_INVALIDARG;
}
// Not needed/implemented
return S_FALSE;
}
//-------------------------------------------------------------------------------------------------
HRESULT STDMETHODCALLTYPE CEvaluatorExample03::GetNormalData
//-------------------------------------------------------------------------------------------------
(
IDreamInputParViewer* pInputPar // Interface to DREAM input parameters
, IDreamDataNormal* pINormalData // Interface to normal distribution data
)
///////////////////////////////////////////////////////////////////////////////////////////////////
{
// Always check pINormalData pointer
if(pINormalData == nullptr)
{
return E_INVALIDARG;
}
size_t d = (size_t)pInputPar->GetProblemDimension();
// Fill mu data
for(size_t iIndex = 0; iIndex < d; iIndex++)
{
pINormalData->AddMeanItem(0.0);
}
// Covariance matrix diagonal value
pINormalData->SetCovarianceDiagonalValue(5.0);
// Data added successfully
return S_OK;
}
//-------------------------------------------------------------------------------------------------
HRESULT STDMETHODCALLTYPE CEvaluatorExample03::EvaluateProposal
//-------------------------------------------------------------------------------------------------
(
IDreamInputParViewer* pInputPar // Interface to DREAM input parameters
, int iSim // input parameter (i-th simulation / Markov chain)
, IDreamMatrix* x // input matrix (m_NumberOfMarkovChains x m_ProblemDimension)
, IDreamMatrix* res // output matrix (Data_Dimension x m_NumberOfMarkovChains)
, BSTR strWorkingDir // path to the current working directory (for current calculation thread)
, BSTR strModelDataDir // path to the permanent directory with model data
, BSTR strModelBinDir // path to the permanent directory with model executables
)
///////////////////////////////////////////////////////////////////////////////////////////////////
/*
DESCRIPTION:
The purpose of this function is to evaluate the current proposal (i.e. one single step
of IDreamInputParams::GetNumberOfGenerations() steps) for iSim-th Markov chain. It computes
the likelihood (or log-likelihood or the residual vector to be compared with the measurement
- see the technical manual) for iSim-th row of input matrix "x" and returns the result
in output matrix "res".
Parameter iSim is from interval <0, IDreamInputParams::GetNumberOfMarkovChains() - 1>.
If pInputPar->GetVectorization() == eVectorization::eVectYes, then iSim = 0 and all proposals
(for all Markov chains) should be evaluated in one step, i.e. during one call of this function.
RETURN VALUE:
S_OK - Operation successful
S_FALSE - Function not implemented (program will continue)
E_FAIL - Operation failed (program will stop)
*/
///////////////////////////////////////////////////////////////////////////////////////////////////
{
/*
Vectorization:
If vectorization is set to "eVectYes" then EvaluateProposal function is called only once with
iSim = 0 and whole input matrix "x" has to be evaluated in one step to get output matrix res.
If vectorization is set to "eVectYes" then EvaluateProposal function is called "x->_colCount"-th
times with iSim = <0, x->_colCount-1> and input matrix "x" has to be avaluated for each column
of matrix "x" separately.
*/
// Temporary vector used for calculations
double* tmpx = nullptr;
FunctionsSC::CopyRowFromIDreamMatrix(iSim, tmpx, x);
// Now loop over each component of mixture
double* loglh = nullptr;
FunctionsSC::CreateVector(loglh, m_k);
for(int j = 0; j < m_k; j++)
{
// Calculate log_likelihood of jth component
// log_lh(:,j) = -0.5 * sum (x - mu(j,:)).^2 , 2) + logF(j);
// vector2 = mu(j,:)
double* vector2 = nullptr;
FunctionsSC::Row(j, vector2, m_mu, m_k, x->_colCount);
// mat1 = x - mu(j,:)
double* mat1 = nullptr;
FunctionsSC::VectorVectorSubtraction(mat1, tmpx, x->_colCount, vector2, x->_colCount);
// mat1 = (x - mu(j,:)).^2
FunctionsSC::VectorElementPow(2.0, mat1, x->_colCount);
// log_lh(j) = -0.5 * sum(mat1) + m_logF(j);
loglh[j] = -0.5 * FunctionsSC::VectorElementSum(mat1, x->_colCount) + m_logF[j];
FunctionsSC::DeleteVector(vector2);
FunctionsSC::DeleteVector(mat1);
}
// Now determine the overall log-likelihood
double maxll = FunctionsSC::VectorMaxElement(loglh, m_k);
// Minus maxll to avoid underflow
double* vector1 = nullptr;
FunctionsSC::CreateVector(vector1, m_k, -maxll);
double* post = nullptr;
FunctionsSC::VectorVectorAddition(post, loglh, m_k, vector1, m_k);
FunctionsSC::VectorElementExp(post, m_k);
// Density(i) is \sum_j \alpha_j P(x_i| \theta_j)/ exp(maxll(i))
double density = FunctionsSC::VectorElementSum(post, m_k);
// Calculate log-L
res->_data[iSim] = log(density) + maxll;
FunctionsSC::DeleteVector(tmpx);
FunctionsSC::DeleteVector(loglh);
FunctionsSC::DeleteVector(vector1);
FunctionsSC::DeleteVector(post);
return S_OK;
}